How can we use the power of crowd, open source, and open innovation to help scientists from various disciplines generate original solutions and advance our current understanding of cancer? Emerging methods in data analysis can create a better understanding of complex fields like medicine or life sciences. But this requires us to phrase challenges in a way that can be comprehendible for doctors, patients, and data scientists alike. The C-K (Concept-Knowledge) method provides a strategy.
Open source publications, communications technologies, and digital platforms have contributed to increased accessibility to knowledge. Scientists are able to be even more open, inviting participants that have expert knowledge and special abilities to solve significant questions.
Access to information, traditionally considered a strategic feature in any scientific research organization, is no longer limited to a few players capable of investing in expensive and lengthy R&D programs. Accurate information is now easily, quickly accessible to almost anyone who is willing to learn, experiment, and make a scientific contribution. Increasing disclosure in scientific processes creates communities in support of scientific projects. Leveraging these open communities to solve various problems and even to create scientific discoveries is becoming increasingly popular.
As Sauermann and Franzoni (2015) point out, a growing amount of scientific research is done in an open manner. Examples can be found across different domains and disciplines.
For instance, the Polymath project, launched in 2009 by Tim Gower, brought mathematicians together to solve important and difficult mathematical problems by creating a common platform where specialists could communicate with one another to find the best route to the solution. Polymath has launched more than 12 challenges. The project proved the benefit of having many minds working together to solve difficult mathematical problems. Harvard, TopCoder, Broad Institute, and the Crowd Innovation Lab launched a joint project to organize a series of challenges to develop algorithms for faster DNA sequence alignment and to improve analysis of gene expression data. These examples clearly demonstrate that open initiatives can deal with extremely complex problems.
Projects like the Polymath Project and the gene expression partnership are often refer to crowdsourced data or open science. They are characterized by open participation, sharing data, and attempting problem solving techniques with participants. Open science promoters often highlight the potential to learn, collaborate with others, and test new theories.
Open science for cancer research
In recent years, life science and medicine have faced major changes with the appearance of new massive sources of information, such as genetic identity and the global patent environment. In parallel, new forms of treatment are becoming more available, like biotherapies and personalized treatments based on patient’s genetic information. Many areas, such as epidemiology, are undergoing major transformations regarding data access and availability that require new methods of data analysis. These disciplines are now using open collaborative settings to explore new ways of dealing with these massive sources of data.
Epidemium, a collaborative initiative to explore new paths for cancer research, was launched in 2016. An inclusive and community-based open science program, Epidemium is a joint program with a pharmaceutical company, Roche, and a community laboratory, La Paillasse. The program uses data challenges, like the Challenge4Cancer, to address the epidemiology of cancer in an open science framework.
Launched in 2016, the first Epidemium challenge was a blast. 678 people participated, creating a broad community of experts with diverse competencies of data analysis, such as statistics, visualization, data mining, oncology, epidemiology. In total, 15 different projects were developed over 6 months. These projects were subjected to evaluation by scientific and ethics committees. The committees judged the scientific validity of the results, ethical implications, level of originality, level of collaboration, potential for impact, and perspectives from patients about the proposed approaches.
From the perspective of knowledge sharing, Challenge4Cancer’s participants had to document their advances and results on a public Wiki page. This transparency allowed for continuous discussion during the challenge and enabled the formation of a vibrant community.
Despite these achievements, some difficulties related to novelty, validity of results, and identification of promising research projects were underlined. One of the critical challenges was the identification of research questions and challenges for future collaboration.
Crowdsourcing and open innovation literature streams emphasize that problem formulation is one of the key factors to ensure successful outcomes and to attract the right participants. Problems should be precise but avoid being too narrow (note Felin & Zenger (2014)). This is particularly relevant for transdisciplinary challenges in open source data analysis. As pointed by Godemann (2008), translating various forms of knowledge from different disciplines is highly beneficial for problem solving, but it creates difficulties in knowledge exchange and integration.
Given the short duration of the Challenge4Cancer and the variety of participants, the appropriation of research topics should be better managed by the community. The research questions should be understandable by the different Epidemium communities, such as doctors, patients, data scientists, and should incentivize them to work together. They also should be original and ambitious so as to attract high-skilled participants.
Given the importance of designing research directions, in 2017 Epidemium decided to launch a preliminary exploration to create a better understanding of the stakes and to identify research questions to tackle.
Shaping new research directions: how design-driven frameworks like Concept-Knowledge can help
Solving questions using new approaches is exciting. But it is crucial to solve the right questions. What is the right knowledge gap to analyze? How do we identify the gaps in cancer research that can be relevant to tackle using data analysis? How do we ensure that the relevant data is collected?
To design research questions, one would normally analyze the existing knowledge gaps and try to formulate questions that are novel enough. In the case of Epidemium, the state of the art is quite broad since it includes disciplines related both to cancer and data analysis. Following traditional literature review would have been too costly and time consuming. Moreover, since the challenge aims to develop entirely new connections between different disciplines, knowledge advances should be presented in a way concise and simple enough to allow non-experts to have a quick understanding of what is going on.
In order to explore the possibilities related to data analysis & cancer research in a systematic way, to identify the framework of the current approaches and to generate a set of innovative concepts, a design theory based framework was applied.
This framework used is based on a design tool derived from the Concept Knowledge (C-K) design theory of innovative design reasoning. We chose design theory since it allows for knowledge expandability that goes beyond pure combinatorial strategies and considers dynamic transformations, adaptations, hybridizations, discovery, invention and renewal of objects discovery. The C-K design framework is useful for understanding novelty since it not only separates state of the art (the available knowledge) and an exploration phase (the concept development), but also defines how to use the existing knowledge to structure the unknown.
C-K Design Theory is based on two interdependent spaces. The Concepts space has a tree-based structure. This tree underlines the design paths for each idea and emphasizes their relations to other fields. The Knowledge space is represented by knowledge databases where different types of knowledge (with mention of its robustness and maturity) can be emphasized.
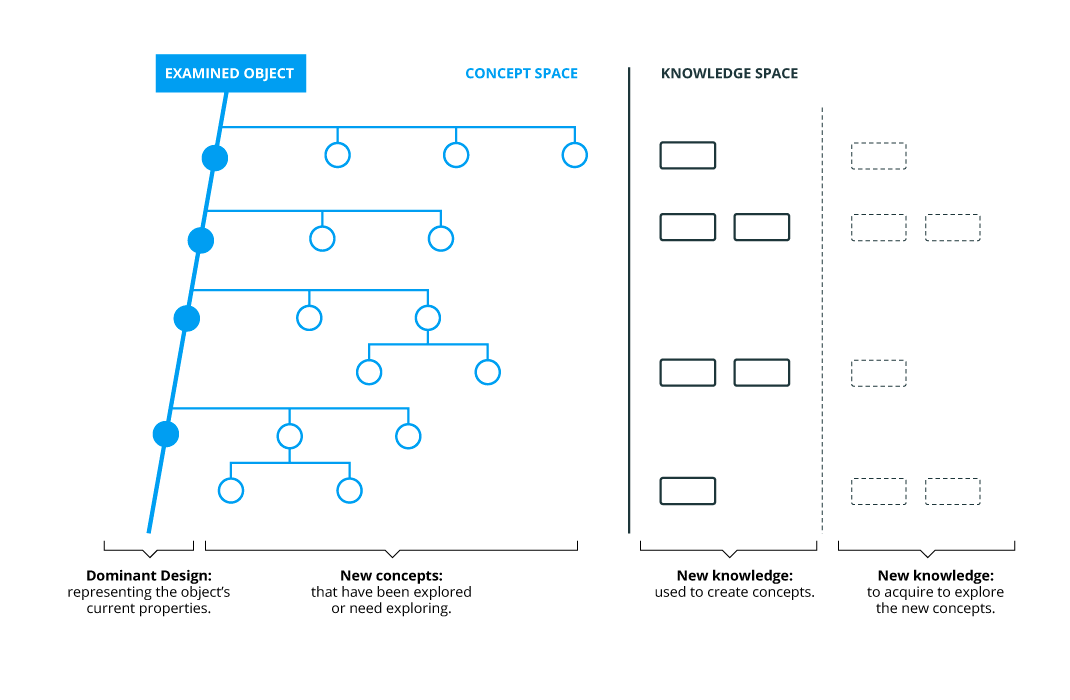
Mapping potential research directions for treating cancer using big data
Along with workshops involving doctors, patients and data scientists, STIM and Mines ParisTech used the C-K design framework to establish a common understanding of cancer and cancer treatment as well as the available data and data analysis techniques that can be used. This step was crucial to build a common vocabulary across experts from different domains, contextualize current approaches to using the C-K framework and define the limits of current approaches.
Once this understanding was made explicit, alternatives were easier to identify by seeking the external knowledge and mapping the existing products. To imagine these alternatives, several workshops were organized with specialists of data analysis and cancer. Literature reviews were completed through close work with the Epidemium team. In total, 25 experts participated in the workshops. They first shared their common vision of the field (i.e., cancer data is used by medical professionals who collect this data and use it to better understand cancer). See Figure 2 for the extract of the map.
Alternatives were proposed at each level of the map. For example, non-experts can use the data, different actors can access the data (and not just medical professionals), and this data can be used differently. Establishing the common understanding helped experts to identify alternatives. For instance, today cancer screening is mostly performed by medical staff. Workshop participants proposed self-screening techniques or screening performed by the third parties when the screening techniques were non-invasive. Moreover, screening should occur not just when the first symptoms appear but on a regular basis. People at risk should be identified (through genome analysis, age, sex, exposure to different risk factors) and they should benefit from frequent individual screening. In the future, continuous screening in real time should even be considered through devices or other monitoring options.
What about data? Different information was relevant depending on the data use, such as data related to the patient health status, to the treatment efficiency and non-efficiency, to the patient’s behavior (nutrition, activity, work), to the environment or other external factors that can affect a personm epigenome data, and data related to patient care services, to the country economy, etc.
Different alternatives were explored and structured thanks to the C-K framework and enabled workshop leaders to identify 45 exploration axes, such as automatically assigning patients to different departments based on factors like type of cancer, socioeconomic status, treatments, assessing treatment efficiency, failure ex post including risk & environmental data, anticipating the efficiency of treatment and side effects per the patient profile and, for each organ, understanding which type of cancer can occur.
The first results were exposed to a larger Epidemium community (around 100 people) for their comments and suggestions. The results were validated by the scientific and ethical committee of the Epidemium community.
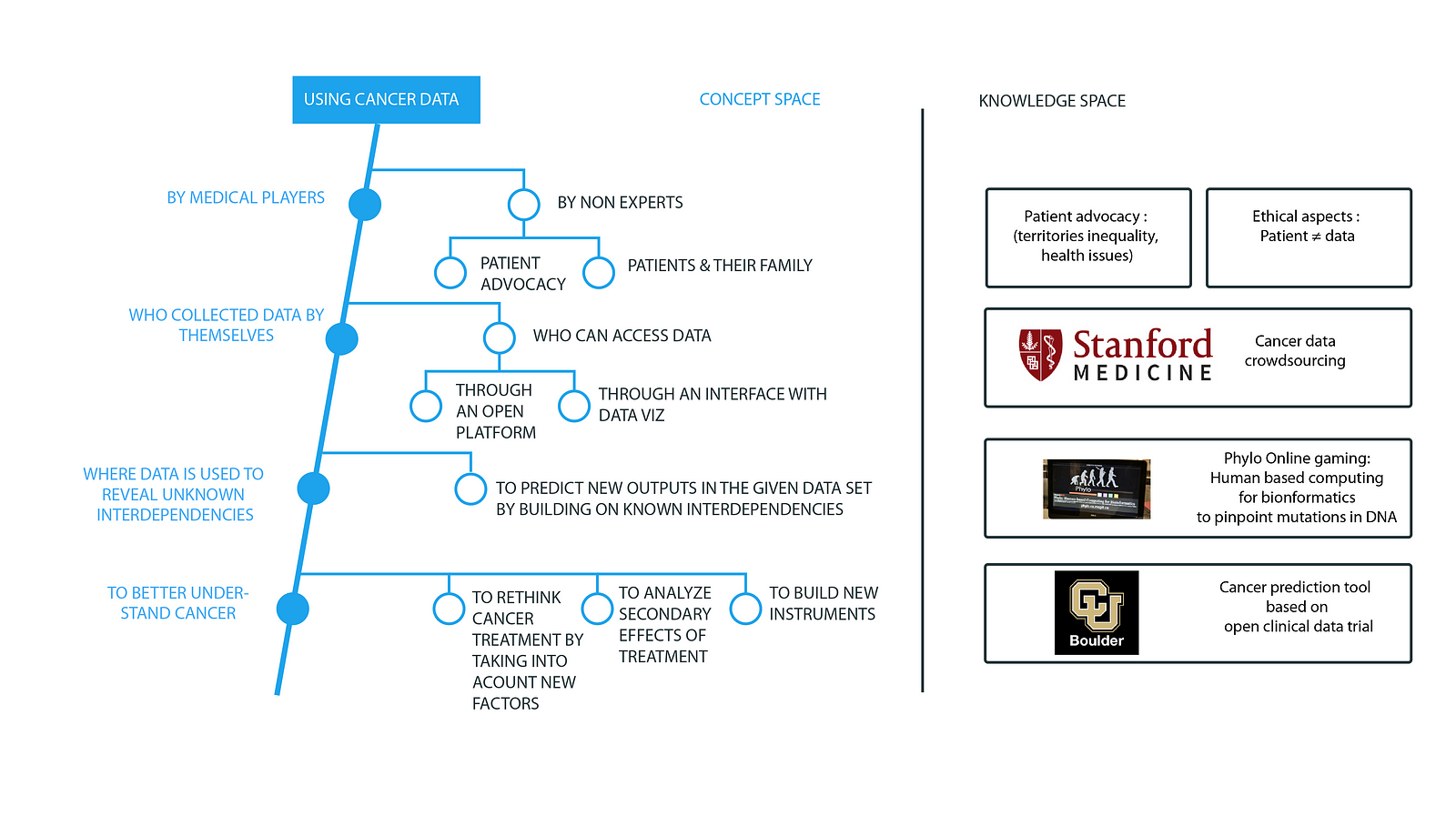
This collaborative work helped the community shape a variety of research directions and identify the knowledge needed to go further. The map is available to anyone who wishes to better understand cancer and its treatment options or who wishes to contribute to the map or complete proposed projects.
Dealing with emerging research directions in a transdisciplinary context
Creating interdependencies between previously unrelated fields or concepts can lead to unexpected ideas. Forcing researchers from different backgrounds to create an interconnected map of concepts related to several rather independent fields allowed the Epidemium community to forge collaboration between different experts and extend the exploration space to create a common understanding related to cancer.
Using design driven frameworks like Concept-Knowledge helped workshop participants to understand and explore various alternatives that Epidemium can use to build research directions and see how other initiatives are positioned, resulting in a visualization of current research on data analysis initiatives for cancer. This map helped explore and generate potential hypotheses that are accessible to the community.
The proposed map is not exhaustive and it is subject to constant changes and improvement. Nevertheless, it offers a comprehensive overview of a complex problem and provides a rich set of research directions.
This approach aimed to provide a systematic exploration of all the possible alternatives and to avoid the cognitive biases that limit participants’ exploration capacity. Moreover, dealing with existing knowledge fostered a better understanding of what is the current state-of-the-art and helped organize the search for new knowledge. It increased the ability of designers to generate original concepts.
We believe that this approach has a potential for developing more general models. It might be interesting to combine the design driven strategy with visualization, text mining, or statistical approaches.
Article co-authored by Olga Kokshagina, STIM and Yohann Sitruk, Mines ParisTech. This article was originally published in Paris Innovation Review on October 13, 2017.
Photo by Louis Reed on Unsplash.



